-
Biochemistry
We are using protein biochemistry (e.g., enzymatic assays, reconstitution) and the powerful Xenopus laevis egg extract system to understand the intricate interactions and regulations of the enzymes involved in cell cycle progression.
-
Quantitative Cell Biology
We are developing and employing quantitative read-outs (e.g., fluorescence cell cycle reporters, single droplet and cell microscopy) in vivo and in vitro to follow the complex dynamics associated with cells undergoing cell division.
-
Computational Biology
We are deriving computational models to account for the vast amount of experimental literature and our own findings. This allows us to derive new hypotheses, identify gaps in our knowledge and gain a deeper mechanistic understanding of cell cycle dynamics.
(Upper panel) Tubulin dynamics in cycling Xenopus egg extracts encapsulated in fluorosurfactant droplets. Metaphase and anaphase spindles are clearly visible. (Lower panel) A novel CDK1 biosensor allows us to monitor the cell cycle dynamics over time in single droplets.
Reconstitution of cell cycle dynamics in vitro
The early embryonic divisions in many organisms, including the African clawed frog Xenopus laevis, are very fast and highly robust - almost clock-like. Despite our detailed knowledge about most of the key players involved in this process, we still do not understand how these rapid cell divisions and all accompanying processes can be executed with such temporal precision.
To answer this question we aim to reconstitute the eukaryotic cell cycle both in vitro and in the frog (Xenopus laevis) egg extract, which amazingly retains the capacity to undergo the biochemical changes associated with cell cycle progression ex vivo. By encapsulating the egg extract into water-in-oil droplets in the presence of fluorescent sensors (which we are developing) allows us to unravel the biochemistry driving this highly robust and precise dynamics.
(Upper panel) Schematic depiction of some of the key regulators of cell cycle progression and their complex interactions that result in the phosphorylation and dephosphorylation of hundreds of proteins during mitosis. (Lower panel) Many mitotic proteins including APC3 (a subunit of the APC/C) and Nup53 (a nuclear pore complex component) are heavily phosphorylated during mitosis and meiosis.
The regulation of multisite phosphorylation during cell cycle progression
We are interested in how the hundreds -if not thousands - of phosphorylation events mediated by the cyclin B-Cdk1 complex, one of the major mitotic kinases, translate into the distinct order of mitotic events necessary for error-free cell division. Particularly, we are interested in proteins and protein complexes, which are phosphorylated at numerous sites raising the question, how the different phosphorylation events are getting integrated to drive function.
To answer this question we combine biochemical assays using frog egg extracts with quantitative mass spectrometry. This approach will reveal new insights into how, when and in which order proteins become phosphorylated during mitosis and how these phosphorylations regulate protein function.
Cell cycle regulation by phosphatases
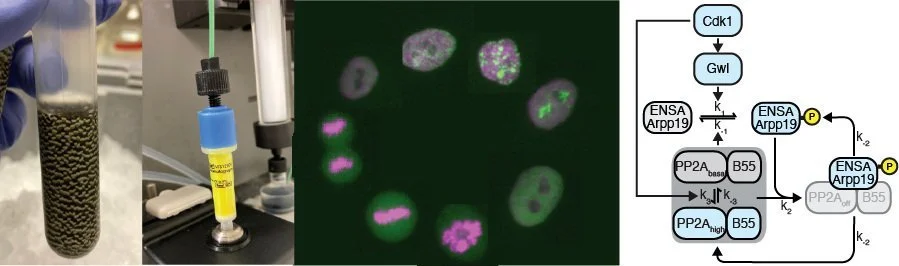
For a long time, phosphatases have been considered to be the unspecific and unregulated counterparts to the sophisticated regulation of cell cycle kinases. However, this picture has dramatically changed, demonstrating that phosphatases are highly specific and highly regulated throughout the cell cycle. We are particularly interested in how signaling pathways directly modulate phosphatase activity in order to regulate cell cycle progression. Currently, we are focusing on the phosphatase PP2A-B55, which we recently showed is both inhibited by Cdk1 through double-negative feedback but also also activated by an unknown Cdk1-dependent mechanism constituting a incoherent feedforward loop (Kamenz et al., 2021).
Using both, biochemical approaches employing frog egg extracts as well as live cell imaging of human cells in cell culture, we will study how Cdk1 activity and cell cycle signaling in general impinge on the activity of PP2A-B55 and other phosphatases.
(Upper panel) Depiction of the regulation of PP2A-B55 by Cdk1 through double negative feedback via GWL/ENSA/ARPP19 and an uncharacterized Cdk1-dependent activation mechanism. (Lower panel) Time course of enzymatic activities of Cdk1 and PP2A-B55 and protein phosphorylation level of a mitotic substrate (Nup53) in a cycling frog egg extract. Note that PP2A-B55 (orange) activity first increases before then decreasing during mitosis.



